NetCDF write performance: Chunking, slicing, openmp
There are multiple ways to improve IO performance of computational models. In some recent experiments I was running I noticed that the write portion of my simulations were taking something like 90% of my runtime. Clearly the simplest way to reduce write times are to reduce the amount of data that is being written, but this was not a reasonable approach to solve my issues. So, instead I explored some options that could help me. This post will focus on using Fortran and NetCDF, and explore three different ways that you might be able to improve write speeds.
The first, is chunking, which is natively implemented in NetCDF. Basically, the idea of chunking is that the NetCDF library will handle some of the complexity for you and fill up a cache before writing output to disk. The second, slicing, is similar to chunking, but places it in the hands of the user. Slicing lets you put multiple values into the NetCDF file sequentially. Finally, if neither of these options are enough, it might be possible for you to use OpenMP to parallelize your write and run tasks.
To get started with this whole thing, we’ll put together a sample model driver that has a main loop, a runner section (proxy for solving model physics), and a writer section.
For now, we’ll just make the main loop do one iteration.
We will write data out into a variable named x, with chunk size chunk and slice size slice.
The simplified code to do this is as follows:
program chunked_sliced_writeout
use testmodule
use netcdf
implicit none
integer :: number_mainloop = 1
integer :: i, n, chunk, slice, narg
real, dimension(:), allocatable :: x
call process_args(n, chunk, slice)
do i=1,number_mainloop
call runner(x, n)
call writer(x, n, slice, chunk)
end do
end programSimple enough, right? The interior code in testmodule is equally simple.
subroutine runner(x, n)
implicit none
real, dimension(:), allocatable, intent(out) :: x
integer, intent(in) :: n
! proxy for doing some complex work
allocate(x(n))
x(:) = 1.0
call sleep(1)
end subroutine
subroutine writer(x, n, slice, chunk)
use netcdf
implicit none
real, dimension(:), allocatable, intent(in) :: x
integer, intent(in) :: slice
integer, intent(in) :: chunk
integer, intent(in) :: n
real, dimension(:), allocatable :: y
integer :: i
character (len=*), parameter :: FILENAME = 'testfile.nc'
integer :: ncid, var_id_x, var_id_y, dim_id_x, dim_id_y, retval
integer, allocatable :: seed(:)
integer size
call error(nf90_create(FILENAME, NF90_NETCDF4, ncid))
call error(nf90_def_dim(ncid, 'x', n, dim_id_x))
call error(nf90_def_dim(ncid, 'y', NF90_UNLIMITED, dim_id_y))
call error(nf90_def_var(ncid, 'xdata', NF90_REAL, dim_id_x, var_id_x))
call error(nf90_def_var(ncid, 'ydata', NF90_REAL, dim_id_y, var_id_y))
call error(nf90_def_var_chunking(ncid, var_id_x, 0, (/ chunk /)))
call error(nf90_def_var_chunking(ncid, var_id_y, 0, (/ chunk /)))
call error(nf90_enddef(ncid))
allocate(y(n))
y(:) = 1.0
do i=1,n,slice
call error(nf90_put_var(ncid, var_id_x, (/ x(i:i+slice) /), &
start=(/ i /), count=(/ slice /)))
call error(nf90_put_var(ncid, var_id_y, (/ y(i:i+slice) /), &
start=(/ i /), count=(/ slice /)))
end do
call error(nf90_close(ncid))
end subroutineSo, with the basics of the code out of the way, we can start some tests.
For all tests I’ll choose a small/moderate array size of 300,000 data points.
As a baseline, I ran with no chunking, slicing, or parallelization.
That is, we’re telling the NetCDF library to put one piece of data into NetCDF at a time.
In my situation it took roughly 27.5 seconds on my laptop to run this (note the one second sleep that I put into the runner subroutine).
The effect of chunking
Since chunking is the easiest to implement I chose to explore it first.
I ran the same code with a chunksize of 10, 100, and 1000.
Generally, in your code to make this happen you just have to ensure that you set the nf90_def_var_chunking after defining the output variables.
This is seen in the writer code above for both the xdata and ydata.
No matter the chunksize this takes roughly 13-14 seconds for my laptop.
That’s almost a 50% reduction in runtime, not bad!
The effect of slicing
Now to see how the slicing affects output, I reset the chunking to 1 and run with slices of 10, 100, and 1000 steps for both variables.
To account for the slices I set the iteration of the inner loop inside of the runner subroutine to step in increments of slice.
Then, I set the start and count arguments of the nf90_put_var subroutine accordingly.
This get’s us to 10.5 seconds or less, even better than just setting the chunksize.
Why not both?
Of course, the next question is, what if we both chunk and slice? As before we wil test chunksizes of 10, 100, and 1000 as well as slice sizes of 10, 100, and 1000. Instead of summarizing the runtimes in text, it seems more reasonable to summarize all of these runs with some plots. The resulting runtimes of each of these is summarized below. But, before I show those results, it is probably worthwile to see one more approach to cutting down on runime.
Enter openmp
OpenMP is a really nice thread-based approach to parallelism that’s built into most modern Fortran compilers.
We can leverage it in this sort of scenario to perform write and run tasks on separate threads.
Of course, in the real world it’s significantly more complex to organize these tasks in separate ways, but hopefully this post can show an abstraction that makes this approach reasonable.
Also, it’s pretty crucial to set up shared memory in an intelligent way so that tons of data-shuffling doesn’t end up outweighing any performance gains on writing output in a seperate thread.
To make this work we will include a few openmp “directives” into the model driver.
Further, instead of complicating tasks, I will just increase the mainloop variable to occur several times.
The modifications to the original driver end up producing this:
program chunked_sliced_writeout
use testmodule
use omp_lib
use netcdf
implicit none
integer :: number_mainloop = 3
integer :: i, n, chunk, slice, narg
real, dimension(:), allocatable :: x
call process_args(n, chunk, slice)
do i=1,number_mainloop
!$OMP PARALLEL SECTIONS
!$OMP SECTION
call runner(x, n)
!$OMP SECTION
call writer(x, n, slice, chunk)
!$OMP END PARALLEL SECTIONS
end do
end programAgain, simpler than you might expect.
The two key portions to all of this working are the two OMP SECTION declarations.
Sections in OpenMP declare specific tasks that can be run in parallel, but only each one at a time.
Note, the thing that we’re trying to reduce here is the effect of having to wait for each subsequent runner subroutine wait on the end of the previous writer subroutine.
That is, after the initial write task is kicked off on it’s thread, the next run task should be free to continue calculations.
Once the run task is complete it should wait for the previous write task to finish.
In essence, only one write and one run task should be occuring at a given moment, where the write task is from an earlier run task.
Since we only have two tasks, running and writing, this sort of program structure is suited to 2 threads only.
It’s pretty simple to set this by running export OMP_NUM_THREADS=2 on Linux (and I assume similarly on MacOS).
If you’re using Windows, you might try getting set up on WSL to try these tasks if it proves difficult natively.
Or just install Linux and embrace the freedom ;)
Bringing it all together
Great, now with all possible scenarios outlined, what is the best overall performance that we can achieve?
To keep things in line, I will report the time per mainloop execution.
The summary of runtimes for each experiment is shown in figure 1, below.
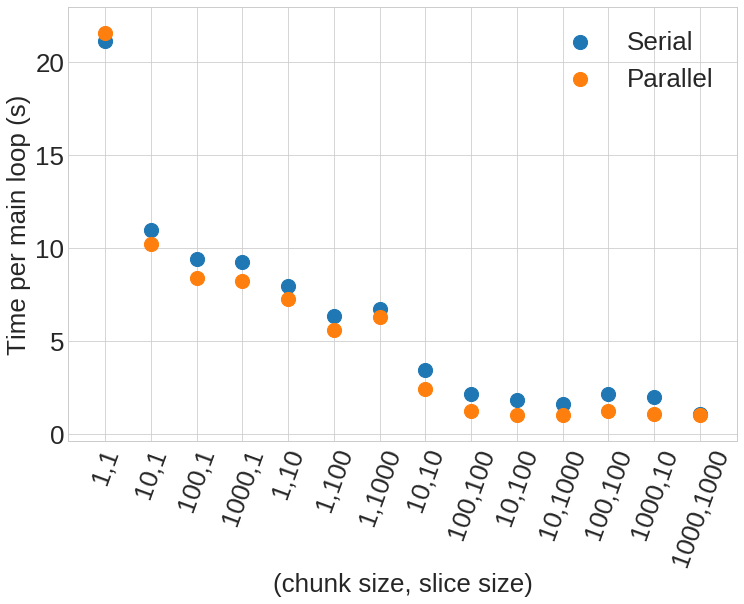
It’s clear from this figure that the no chunk, no slice run (first column) is significantly worse than any of the other runs.
So, to first order, these experiments insinuate that taking the naive approach may hurt your scalability quite easily.
The next 6 columns show the results from simply implementing chunking or slicing, but not both.
This nets a runtime performance boost of 2-4x which is quite nice, especially in the case of chunking since it requires no modification of your IO code, but merely an extra call to nf90_def_var_chunking.
However, if it’s possible to implement both chunking and slicing even further benefits can be reaped.
This is shown in columns 8-14 of figure 1, where runtimes can decrease to the point that the IO portion of the run is negligible.
In the case of using chunking and slicing sizes of 1000 each the runtime devotes <5% to IO, since I have hard coded a 1 second sleep into the runner subroutine call as a proxy for doing “more complex computation”.
Clearly, if this level of IO performance can be acheived, the need for OpenMP is reduced.
However, some of the other experiments show that when IO is still on the same order of runtime as the computation, adding OpenMP can decrease overall runtimes.
Columns 2-13 of figure 1 show this where the runtime of the parallel version of the code reduces runtime on average by about 1 second (that is, the time I hard-coded as “more complex computation”)
Godspeed all ye who try to implement this
Despite all the rather nice results for a simple and contrived test case I must lament that actually changing the code of more complex models to maximize this performance can be a trying task. When writing numerical code the IO performance is something that’s often not considered as a first order determining factor for overall performance, and in many cases that is a fair assumption. But, when that assumption breaks down it often does so in a pernicious way and is not easily fixed by simple (or even savvy) users of the code.